Molecular cloning and analysis of lymphokines / edited by David R. Webb, David V. Goeddel.
- Date:
- 1987
Licence: Attribution-NonCommercial 4.0 International (CC BY-NC 4.0)
Credit: Molecular cloning and analysis of lymphokines / edited by David R. Webb, David V. Goeddel. Source: Wellcome Collection.
44/344 (page 26)
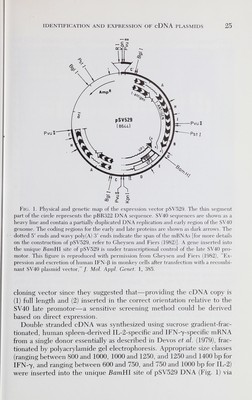
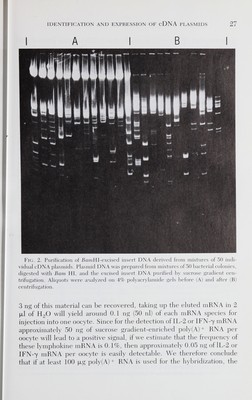
![26 rene devos et al. G/C homopolymer tailing and used to transform E. coli (strain DHl for IFN-7, and strain HBIOI for IL-2). Between 5000 and 10,000 colonies were obtained for each size group. Plasmid DNA was prepared from mixtures of 50 individual clones from the IFN-7 cDNA library and used to transfect AP8 monkey cells (Gheysen and Fiers, 1982). After testing 8000 clones (160 groups of 50 colonies), not one group of 50 clones led to a significant IFN activity in the supernatant of the transfected cells. We concluded that either no full length IFN-7 cDNA copy, inserted in the correct orientation, was present in our library or IFN-7 was expressed below the detection limit of our assay. Later (Devos et al., 1982a), we showed that our cDNA library indeed contained a IFN-7 cDNA clone which resulted in the appearance of IFN-7 activity after transfection of AP8 cells, but that the level of ex¬ pression was about 100 times lower than the IFN activity of IFN-ß produced after transfection of AP8 cells under similar conditions with a homologous IFN-ß cDNA containing plasmid. Our inability to identify an IFN-7 cDNA clone using this direct-screening method led us to use cDNA-mRNA hybridization followed by translation in oocytes to screen our IFN-7 and IL-2 cDNA libraries. Human spleens proved to be a reliable source of both IFN-7 and IL-2 mRNA necessary for the suc¬ cessful identification of both IFN-7- and IL-2-cDNA clones. B. Preparation of cDNA Inserts For the identification of specific cDNA clones by mRNA selection, groups of individual recombinant plasmids are immobilized on nitro¬ cellulose filters, hybridized with mRNA and bound mRNA, eluted, and translated in vitro (Derynck et al., 1980; Parnés et al., 1981). To increase the hybridization efficiency, and thus the potential signal after transla¬ tion, we decided to immobilize only insert DNA onto the filters instead of binding total plasmid DNA. On the other hand, since the insert DNA comprises 10% of the total plasmid DNA, binding only insert DNA allows the screening of a greater number of different colonies per filter. Insert DNA from mixtures of 50 individual clones was prepared by BamHl digestion (the BamHl site of the vector pSV529 was restored by filling in the 5' protruding ends before the dC homopolymer tailing), purified by sucrose gradient centrifugation (Fig. 2), and bound on nitro¬ cellulose filters (Kafatos et al, 1979). We estimated that at least 10 inserts were present per group of 50 colonies and that approximately 5 fxg insert DNA per group or 0.5 fxg of each insert was immobilized on each filter. Assuming that 10 ng of each mRNA species can hybridize to each individual insert [10% of total poly(A)+ RNA, see below] and that](https://iiif.wellcomecollection.org/image/b18037082_0045.JP2/full/800%2C/0/default.jpg)